Emerging roles of GLIS3 in neonatal diabetes, type 1 and type 2 diabetes
- 1Division of Endocrinology, Department of Medicine, MetroHealth Medical Center, Case Western Reserve University, Cleveland, Ohio, USA
- 2Department of Anesthesiology, The First People’s Hospital of Foshan & Foshan Hospital of Sun Yat-sen University, Guangdong, China
- Correspondence should be addressed to Y Yang; Email: yisheng.yang{at}case.edu
-
Figure 1
Schematic diagrams of mouse and human GLIS3 gene and protein structures. (A) Mouse Glis3 gene and protein structures. The full-length mouse Glis3 gene contains 11 exons and encodes a protein of 935 aa. The ZFD is encoded by part of exon 4 and is responsible for the binding to the consensus GLIS3RE of its target genes such as insulin, Ngn3 and Ccnd2. TAD is localized at its C-terminus. CUL3 and ITCH promote GLIS3 polyubiquitination and degradation via the proteasomal pathway. SUFU, interacting with GLIS3 through the conserved VYGHF motif at N-terminus of GLIS3, was shown to inhibit the association of CUL3 with GLIS3, thereby protecting GLIS3 protein from proteolytic degradation. (B) Human GLIS3 gene and protein structures. Human GLIS3 gene have two major variants that encode GLIS3 protein isoforms a and b that share the last 9 exons. The variant 1 encodes the longest isoform a with 930 aa. The variant 2 harbors a distinct 5′ UTR and lacks an in-frame portion of the 5′ coding region, resulting in a shorter N-terminus in isoform b with 775 aa, compared to variant 1.
-
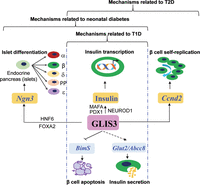
Figure 2
Current understanding of the molecular actions of GLIS3 in diabetes. This figure summarizes the pathways by which ablation, loss-of-function mutations or functional impairment of GLIS3 cause neonatal diabetes, as well as common T1D and T2D. See text for details. A full colour version of this figure is available at http://dx.doi.org/10.1530/JME-16-0232.
- © 2017 Society for Endocrinology