Molecular diversity of corticotropin-releasing hormone mRNA-containing neurons in the hypothalamus
- 1Department of Molecular Neurosciences, Center for Brain Research, Medical University of Vienna, Vienna, Austria
- 2MTA-SE NAP Research Group of Experimental Neuroanatomy and Developmental Biology, Hungarian Academy of Sciences, Budapest, Hungary
- 3Department of Anatomy, Semmelweis University, Budapest, Hungary
- 4Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden
- Correspondence should be addressed to R A Romanov or T Harkany; Email: Roman.Romanov{at}meduniwien.ac.at or Tibor.Harkany{at}meduniwien.ac.at
-
Figure 1
Molecular heterogeneity of Crh+ neurons in the hypothalamus. (A) Distribution of GFP+ neurons outside the PVN in the hypothalamus of Crh-IRES-Cre::GFP mice. (B) Single-cell RNA-seq distinguishes 62 neuronal subclasses along the PVN–arcuate axis of the hypothalamus (Romanov et al. 2015). Expression levels (horizontal axis) were plotted as means of log2 transformed mRNA copy numbers ± s.e.m. Red and green colors identify GABAergic (#14, #15 and #24) and glutamatergic clusters (#44 and #45) of hypothalamic neurons, which express Crh mRNA at levels exceeding 2× the s.e.m. *q < 0.05 (Wilcoxon rank-sum test corrected for multiple testing). (C) Among both GABA and glutamate neurons, subsets are endowed with Crh mRNAs. (D) CRH co-exists in Lhx6-GFP neurons (asterisks) particularly in the bed nucleus of stria terminalis and preoptic area. (E) Neurotransmitter heterogeneity of Crh+ neurons. Venn diagrams demonstrate the proportion of dual and triple neuronal phenotypes in GABA/glutamate (top), GABA/dopamine (middle) and GABA/glutamate/dopamine (bottom) neurons. This analysis was performed using a threshold for mRNA expression at a level of ≥2 mRNA transcripts for each gene. Percentage values indicate the proportion of Crh+ neurons falling into groups and intersections (‘dual phenotype’ categories). Scale bars = 250 µm (A), 10 µm (D). Adapted, with permission, from Romanov et al. (2016).
-
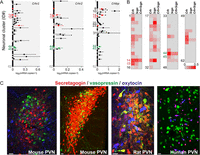
Figure 2
Molecular determinants of CRH signaling in the mouse hypothalamus. (A) Neuronal subclusters expressing CRH receptors (Crhr1 and Crhr2) and CRH-binding protein (Crhbp). Note that some cells in clusters #14 (GABA) and #44 (glutamate) can express at least one receptor and binding protein for CRH signaling. Red and green colors identify respective GABAergic and glutamatergic clusters, which express Crh mRNA at levels exceeding 2× the s.e.m. Note that significant levels of gene expression were found only for Crhr2 and Crhbp but not for Crhr1 within the hypothalamus (*q < 0.05, Wilcoxon rank-sum test corrected for multiple testing). (B) Secretagogin expression among hypothalamic neurons. Cluster #44 (glutamate) coexpresses both Scgn and Crh genes, and this cluster localizes to the PVN (Romanov et al. 2015). (C) Secretagogin expression in the hypothalamus of laboratory rodents and humans. In mouse, secretagogin co-exists with neither oxytocin nor vasopressin. In contrast, a subset of vasopressin+ and oxytocin+ neurons can co-express secretagogin in rat and human, respectively. Color code: secretagogin (red), vasopressin (green) and oxytocin (blue). Scale bars = 20 µm. (A and B) adapted, with permission, from Romanov et al. (2016).
-
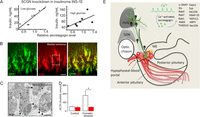
Figure 3
Secretagogin affects hormone secretion. (A) Secretagogin knockdown in INS-1E insulinoma cells limits insulin release. (B and C) Secretagogin co-localizes with CRH at the median eminence (B) and accumulates as a membrane-associated synaptic component at the ultrastructural level (C). (D) Plasma ACTH levels 12 min after formalin (4%) injection into a paw of conscious adult mice. Red color: secretagogin knockdown in vivo. Note that secretagogin deletion occludes ACTH release. *, P < 0.05 vs. formalin-stressed controls. (E) The scheme showing involvement of secretagogin in hypothalamic CRH release, including potential protein-protein interactions. Scale bars = 10 µm (B) and 500 nm (C). Reproduced, with permission, from Romanov et al. (2015).
- © 2017 Society for Endocrinology