Circadian clocks: regulators of endocrine and metabolic rhythms
- (Correspondence should be addressed to M Hastings; Email: mha{at}mrc-lmb.cam.ac.uk)
Abstract
Daily and seasonal rhythms in the endocrine system are co-ordinated by a hypothalamic pacemaker, the suprachiasmatic nuclei (SCN) that is synchronised to solar time by direct retinal afferents. Individual SCN neurons are circadian clocks, their intrinsic oscillator consisting of a series of interlinked autoregulatory transcriptional/post-translational feedback loops incorporating Period (Per) and Cryptochrome (Cry) genes. Mutations that alter the rate of transcription of Per and Cry genes or the stability of Per and Cry proteins affect clock speed. Molecular timekeeping in SCN neurons is synchronised and sustained by interneuronal neuropeptidergic signals. A molecular clock mechanism comparable to that of the SCN is present in most major organ systems. These tissue clocks are synchronised by endocrine, autonomic and behavioural cues that are dependent on the SCN, and in turn they drive the circadian expression of local transcriptomes, thereby co-ordinating circadian metabolism and physiology. Rhythmic glucocorticoid signalling is a prominent mediator of SCN output and internal synchroniser. The role of local SCN-synchronised clocks in controlling vital processes, including xenobiotic detoxification, cell division and nutrient metabolism, is essential to health, and disturbances to circadian timing arising from modern working schedules are becoming recognised as an increasingly relevant factor in major systemic illness. Moreover, the newly identified molecular components of circadian control systems provide novel avenues for therapeutic intervention.
Introduction
The endocrine axis is forever dynamic. The synthesis and secretion of hormones change predictably over the course of hours, months, years and lifetimes (Hastings 1991, Czeisler & Klerman 1999). The most dramatic, recurrent and persistent changes are those that occur over the 24-h solar cycle. As we engage with the world during daytime wakefulness and then withdraw from it during nocturnal sleep, our bodies alternate between catabolic and anabolic states, thereby segregating incompatible metabolisms in time. These global changes are governed by daily cycles of hormonal activity, different endocrine axes preparing for and sustaining temporally precise programmes in cardiovascular activation, cell growth, renal filtration, nutrient mobilisation and xenobiotic metabolism, etc. (Hastings et al. 2003). Indeed, all of the principal physiological axes are subject to daily modulation and endocrine signals, in partnership with the autonomic nervous system, are the major co-ordinators of this internal rhythmicity (Buijs & Kalsbeek 2001). The purpose of this paper is to review recent developments in our understanding of the fundamental mechanisms of the biological clocks that drive endocrine and other daily rhythms, and then to apply this basic knowledge to understand what happens toour health and well-being when the body’s clockwork goes wrong.
The suprachiasmatic nuclei(SCN)asa circadian clock
It is perhaps unsurprising to observe daily rhythms of metabolic function and endocrine activity in subjects who are pursuing their normal lives: after all, the internal changes may merely reflect different patterns of behaviour imposed by the cyclical environment. A critical finding, however, is that when humans or experimental animals are held in temporal isolation, deprived of direct contact with the solar or social world, their daily cycles do not become disorganised or peter out. Rather, they continue with a period of ~1 day, hence circadian (Fig. 1⇓) and these rhythms can persist for weeks, even years, free-running with a high amplitude and exquisite precision (Aschoff 1984, Czeisler & Klerman 1999). This simple result demonstrates the existence of internal circadian clocks: self-sustaining endogenous biological timekeepers. Under natural conditions, these clocks are synchronised (entrained) to the external world, principally via light cycles but also via social routines (Mrosovsky et al. 1989, Pittendrigh 1993). This ensures that they run with a period of exactly 24 h, and so provide an internal representation of solar time phase locked to dawn and dusk. The biological advantage of such entrained clocks is that they enable the individual to anticipate and thereby prepare for the challenges and opportunities of day and night and also seasonal changes. This pre-adaptation makes more efficient biological machines (Woelfle et al. 2004, Dodd et al. 2005), a selective advantage that presumably underlies the independent evolution of circadian clock mechanisms across the major taxa, including cyanobacteria, fungi, higher plants and animals (Dunlap 1999). Moreover, it can explain why the clock has been co-opted to regulate so many physiological processes in humans (Hastings et al. 2003).
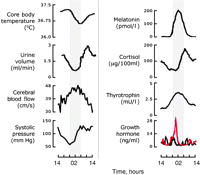
Representative physiological and endocrine circadian cycles in humans held under constant routine conditions. Left panel shows circadian variation in four vital physiological functions, whilst on the right are four endocrine axes subject to circadian control. The shaded area depicts when the subjects would normally have been sleeping but in the constant routine they remained awake. Throughout the routine subjects were held in dim light, deprived of time cues, recumbent and given frequent, regular but limited water and food. In the case of growth hormone, levels are not rhythmic in the absence of sleep (black line) but if sleep is permitted a clear circadian cycle emerges (red line). Figure modified from Czeisler & Klerman (1999), Hastings et al. (2003) and Conroy et al. (2005).
The principal circadian clock in the brain of mammals consists of the paired SCN that sit at the base of the hypothalamus, above the optic chiasm on either side of the third ventricle (Reppert & Weaver 2002). These clusters of ca 10 000 GABAergic neurons are in turn subdivided into a ventral ‘core’ region, which also express vasoactive intestinal polypeptide (VIP) and receive direct innervation from the retina and areas of brain stem that are responsible for entrainment, and a dorsal ‘shell’ region containing arginine vasopressin positive cells, which appears to be the primary pacemaker, its output driving behavioural and other rhythms (Hastings & Herzog 2004). Damage to the SCN can render experimental animals arrhythmic and cause sleep disorders in patients, whilst intracerebral grafts of perinatal SCN can restore behavioural circadian rhythms of SCN-ablated rodents (Weaver 1998). Electrical firing rates, cellular metabolism and gene expression within the SCN all exhibit spontaneous circadian rhythms both in vivo and in isolated culture. Indeed, even individual SCN neurons in dispersed culture are effective circadian timekeepers, as evidenced by their robust cycles of electrical activity and gene expression (Aton et al. 2005, Liu et al. 2007a). In this regard, the SCN clock is a remarkable and very rare piece of neurobiology because the behavioural phenomenon observed at the level of the entire organism, circadian timekeeping, can be seen not only at the level of a tissue but also within individual neurons.
The SCN control endocrine cycles in particular and wider metabolic rhythms in general by two general means. First, via their anatomical connections to centres controlling sleep and wakefulness, the SCN determine the timing of sleep and thereby the timing of sleep-dependent events, for example, the nocturnal secretion of prolactin and growth hormone (Fig. 1⇑; Czeisler & Klerman 1999). Separately, through their connections to the neuroendocrine and autonomic systems (see below), the SCN clocks can drive hormonal and other rhythms independently of sleep; hence these rhythms, e.g. melatonin and cortisol, continue under constant routines where subjects are prevented from sleeping. Many circadian rhythms exhibit intermediate effects, e.g. core body temperature cycles have greater amplitude when subjects sleep but are nevertheless clearly expressed during sleep deprivation.
Circadian clock genes
How might a single neuron be a circadian clock? The past decade has witnessed a remarkable advance in understanding the cellular clockwork, the major breakthroughs being based on studies of circadian mutants, first in fruit flies and then in rodents (Reppert & Weaver 2002, Hastings et al. 2003). The basic model is of an autoregulatory delayed negative feedback loop in which the protein products of particular clock genes negatively regulate their own transcription (Fig. 2⇓). Such an oscillatory model is not unknown to endocrinologists, where negative feedback from target tissues often sustains rhythmic hormone release, but in the circadian system the time scale of the events is drawn out to create a long but nevertheless stable cycle of about 24 h. The central factors in this model are two gene families: Period (Per1, Per2 and Per3) and Cryptochrome (Cry1, Cry2). Transcription of these genes is activated at the beginning of circadian day by heteromeric complexes containing Clock and Bmal1 proteins that act through the E-box regulatory sequences of their target genes. Clock and Bmal1 both contain basic helix–loop–helix (bHLH) motifs for DNA binding at their N-terminus and so-called Per-Arnt-Sim (PAS) domains, first identified in Drosophila Per, which facilitate protein/protein interactions. In addition, Clock carries a glutamine-rich region in the C-terminus and has intrinsic histone acetyltransferase activity, both of which are important for transactivation (Doi et al. 2006). Under this activation, levels of Per and Cry mRNAs accumulate in the SCN over circadian daytime, with rising protein levels lagging behind by several hours. As accumulation of Per and Cry proteins finally peaks in the nuclei of SCN neurons at the end of circadian daytime, mRNA levels start to decline due to the negative feedback actions, particularly of Cry proteins. Although Cry proteins are known to associate with Clock and Bmal1, the molecular details of this feedback suppression are unclear – is their interaction with DNA altered, perhaps by different phosphorylation state (Kondratov et al. 2006, Dardente et al. 2007)? Do Cry proteins physically interpose between Clock and Bmal1 and compromise their dimeric action and how does alternating binding by Clock/Bmal1 and Per/Cry proteins affect histone structure (Etchegaray et al. 2003, Ripperger & Schibler 2006)? The C-terminus of Bmal1 is critical for both transcriptional activation and repression by Cry protein and thus may form part of a switch mechanism (Kiyohara et al. 2006). It is clear that such feedback is essential for the oscillator to run: mutant forms of Clock and Bmal1 that are not susceptible to the negative feedback actions of Cry proteins cannot sustain circadian cycles of gene expression (Sato et al. 2006).
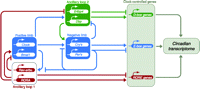
Schematic representation of the feedback loops that make up the molecular oscillator and its principal output pathways for co-ordination of circadian gene expression. The core oscillation (blue lines) is driven by the positive limb of Clock(Npas2)/Bmal1-dependent activation of Per and Cry genes and the subsequent negative feedback by Per/Cry protein complexes onto their encoding genes. These positive and negative actions are mediated through E-box regulatory sequences in the Per and Cry genes. This core is supported by an ancillary loop (red lines) involving a second series of targets, including RevErbαand Rora, which are also expressed rhythmically as Clock/Bmal1 and Per/Cry proteins interact with their E-boxes. RevErbα, Rora and related factors in turn act via RORE sequences in Bmal1 to drive its rhythmic expression in anti-phase to Per and Cry genes. A further output that also feeds back into the core loop involves Dbp/E4bp4 proteins (green lines). This pathway is also rhythmic, Dbp being regulated by E-box and RORE inputs, and it in turn can act via D-box sequences within some of the central genes (including the Per genes and Rev-erbα(data not shown)). This resonant network of circadian gene expression ensures that the various circadian proteins, both positive and negative factors, oscillate within the cell in a precise sequence. Consequently, these circadian proteins will time the expression of downstream cohorts of clock-controlled, output genes that carry E-box, D-box and/or RORE sequences. These primary clock-controlled genes will in turn regulate second-order clock-controlled genes, ultimately orchestrating the circadian transcriptome that underpins local circadian metabolism. Modified from Preitner et al. (2002) and Ueda et al. (2005).
The core loop described above is augmented and stabilised by accessory pathways (Fig. 2⇑), the most obvious involving two orphan nuclear receptor proteins, Rev-Erbαand Rora. Both are activated in phase with the Per and Cry genes by Clock and Bmal1, but in turn theyaffect Bmal1 expression. While Rora has a positive role, RevErbαis a suppressor of Bmal1, and their co-ordinate action through RORE regulatory sequences within the Bmal1 gene suppresses Bmal1 mRNA levels during the day and early night, but as RevErbαdeclines at night, the net result is that Bmal1 is activated (Preitner et al. 2002, Sato et al. 2004). Consequently, Bmal1 and Per mRNAs oscillate in anti-phase, and a surge of Bmal1 expression at the start of circadian day provides an additional ‘boost’to initiate the new cycle of Per and Cry gene expression just as negative feedback is waning – perfect timing.
A consequential feature of these multiple loops is redundancy. Single knockouts of Per1, Per2, Per3, Cry1, Cry2, Rev-Erbαand Clock may alter circadian period but these mutations do not disable the clock (Ko & Takahashi 2006). Deletion of both Per1 and Per2, or both Cry1 and Cry2 is required to stop circadian cycling. Only Bmal1 appears to be the single essential factor in the network, although a mutation of Clock that impairs its transactivational role lengthens and destabilises circadian period. In the complete absence of Clock, however, circadian competence within the SCN is maintained by a closely related protein, Npas2, which can partner Bmal1 (DeBruyne et al. 2007a,b).
Additional factors that are controlled by the core loop and also interact with some of its components include the bHLH proteins Dec1 and Dec2, which can inhibit Clock/Bmal1 activity (Honma et al. 2002), and a pair of transcriptional regulators, Dbp and E4bp4, which can act antagonistically, often through D-box regulatory sequences in their target genes (which include Per1–3 and Rora; Ueda et al. 2005). Although these accessory loops may stabilise the core system, it seems more likely that their primary role is as outputs from the clockwork, as levels of RevErbα, Rora, Dbp and E4bp4 oscillate over time, in and out of phase with Per and Cry proteins, they will direct phase-specific downstream transcriptional cascades. Such temporally defined regulation of target genes carrying functional E-boxes, D-boxes and ROREs will ultimately choreograph the circadian life of the SCN cell. Indeed, transcriptional profiling by DNA microarray studies suggests that several hundred SCN genes are under phase-specific clock regulation and that they control key neuronal processes such as energy metabolism and vesicle recycling that underpin circadian cycles of electrical and neurosecretory activity (Panda et al. 2002, Ueda et al. 2002).
Setting clock speed
To be adaptive, circadian period approximates 24 h, but how is this achieved? A complete understanding of the molecular dynamics within the clock is not yet available but it is clear that certain key processes are tuned to generate a 24-h cycle. The most obvious is the rate of transcription of the Per and Cry genes which dominates circadian day: as noted above, the Clockdelta19 mutation, which compromises transcriptional activation, lengthens circadian period in mice to ca. 28 h. Protein stability is another key factor (Gallego & Virshup 2007). In the tau mutant hamster, dysfunctional activity of casein kinase 1ε(CK1ε) alters the phosphorylation of Per proteins, leading to their accelerated proteasomal degradation (Lowrey et al. 2000) and clearance from the nuclei of SCN neurons (Dey et al. 2005). Homozygous hamsters consequently have a 20-h circadian period in behaviour and endocrine profiles, including melatonin, cortisol and luteinising hormone (Lucas et al. 1999). Intriguingly, a mutation of human CK1δ which is closely related to CK1ε, is similarly linked to accelerated period and advanced sleep (Xu et al. 2005), whilst a mutation of Per2 in humans, which also affects it phosphorylation state, similarly accelerates circadian period, and is manifest by an extreme sleep disorder of familial advanced sleep phase syndrome (FASPS; Toh et al. 2001). The biochemical basis to these fast clocks remains unsolved, however (Vanselow et al. 2006). Mice expressing hPER2 mutations do show accelerated period, confirming the contribution of Per2 to setting clock speed, but the underlying phosphorylation of Per2 in these mice is complex and the relative contributions of CK1δ, CK1ε and other kinases are unclear (Xu et al. 2007). It is likely that some kinases are responsible for a limited number of priming phosphorylations at particular Per2 residues (including the FASPS site), which then trigger serial phosphorylations by other kinases (possibly CK1ε) at many other sites. Moreover, the functional consequences of these modifications to Per2, particularly how they relate to decreased stability, await clarification.
Once appropriately phosphorylated, proteins destined for degradation by the proteasomal pathway are poly ubiquitinylated and several circadian-relevant ubiquitin ligases have been identified in lower organisms. In mammals, Per proteins are thought to be drawn to the ubiquitin ligase complex by the F-box protein βTrcp, although the effects of βTrcp mutations on circadian timing in vivo await clarification. In contrast, it has been demonstrated both in vivo by mutant mice and in vitro by biochemical analyses that Cry proteins are targets for Fbxl3 (Busino et al. 2007, Godinho et al. 2007, Siepka et al. 2007). This F-box protein binds to the ubiquitin ligase complex by the N-terminal F-box and recognises its targets through its leucine-rich repeat region (LRR). Mutations in the LRR reduce affinity for Cry proteins, thereby slowing down their clearance from the cell. This prolongs the duration of the phase of negative feedback within the clock, leading in turn to reduced expression of Per1 and Per2 mRNAs and proteins and a marked prolongation of circadian period by 3 h. This period lengthening represents a selective extension of late circadian night as the onset of a new cycle of circadian gene expression is delayed by elevated Cry.
Entrainment of the SCN by light
Tau mutant hamsters, Clock mutant, hPer2 mutant and Fbxl3 mutant mice show atypical patterns of synchronisation to 12 h light:12 h darkness cycles; characteristically, the short-period mutants become active during the light phase, whereas activity onset in long-period mutants is delayed some hours after lights off. Wild-type mice and hamsters typically become active at lights off, of course, but the extremes exhibited by the mutants (and FASPS patients) are an exaggeration of the normal range of entrainment patterns that in humans are colloquially known as ‘larks’ and ‘owls’ and more formally as chronotypes (Zavada et al. 2005). The phase of entrainment reflects a balance between intrinsic circadian period (and its difference from 24 h) and the resetting effect of daily light schedules. In both nocturnal rodents and humans, light experienced around dusk and dawn is particularly relevant, causing phase delays and advances to the SCN clock respectively (Khalsa et al. 2003). Remarkably, conventional rod- and cone-based photoreceptors in the retina are not necessary for circadian entrainment. Rather, a novel photopigment, melanopsin, which was first discovered in amphibian skin, is active within a sub-population of intrinsically photoreceptive retinal ganglion cells (Qiu et al. 2005). This pathway alone, via its direct projections to the SCN, is sufficient to entrain the clockwork, and entrainment is lost completely in mice lacking melanopsin, rods and cones (Hattar et al. 2003, Panda et al. 2003).
Within the SCN core, glutamatergic retinal terminals from melanopsin cells trigger a calcium-dependent signalling cascade involving MAP kinases that ultimately activates, by phosphorylation, calcium/cyclic AMP response element (CRE)-binding protein (CREB). This response is clockgated, insofar as light delivered when cellular activity in the SCN is high during circadian day is ineffective, whereas light delivered in circadian night when the SCN are quiescent triggers the cascade. Genes that carry CREs, including Per1 and Per2, are rapidly induced by light in the core SCN, and the electrical firing of these neurons is increased for several hours. This in turns leads to trans-synaptic activation of SCN shell neurons, likely via release of GABA, VIP and GRP from the core neurons, and further induction of Per1 and Per2 expression (Hastings & Herzog 2004). The consequences for the clockwork of this induction of Per genes depend on when it happens. At dusk/early night, spontaneous expression of the genes is falling and so induction will acutely delay the oscillator. Conversely, during late circadian night/dawn, levels are low and starting to increase and so induction will accelerate these spontaneous changes and hence advance the oscillator. This model therefore provides a molecular explanation for the differential resetting actions of dusk (delay) and dawn (advance) light on circadian behavioural and physiological rhythms.
Real-time imaging of the SCN circadian circuitry
The identification of Per1 and Per2 as components of the SCN oscillator facilitated a revolutionary way of (literally) viewing the clockwork. Exploiting techniques first applied to the clock of Arabidopsis and Drosophila; transgenic mice and rats were created in which elements of the Per1 promoter were used to drive the expression of either firefly luciferase (Yamazaki et al. 2000, Yamaguchi et al. 2003) or green-fluorescent protein (Kuhlman et al. 2000). This has been extended to a fusion protein reporter, where luciferase coding sequence is tagged onto the Per2 protein (Yoo et al. 2004). Bioluminescence or fluorescence recordings of cultured SCN tissue slices from these animals have revealed the exquisite precision of the molecular events at the heart of the clockwork, with high-amplitude circadian cycles of gene expression that can continue indefinitely across the whole SCN or within individual SCN cells (Fig. 3⇓). Moreover, these SCN oscillations are affected by clock gene mutations in the same way that the mutations affect behavioural rhythms – abolishing them or altering period (Godinho et al. 2007, Liu et al. 2007a).
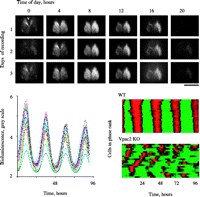
Real-time imaging of SCN circadian gene expression in culture. Top panel presents serial frames from organotypic SCN slice from PER2::LUC reporter mouse recorded in static culture for 3 days and imaged with CCD camera. Note extensive activation and decline of gene expression across SCN with initial increase in dorsomedial region adjacent to third ventricle (v) (bar=1 mm). Lower left panel depicts bioluminescent activityof 25 representative individual SCN cells record from a different slice. Each cell exhibits clear circadian cycles with maintained phase and amplitude. Lower right panel presents raster plots of cellular circadian gene expression from wild-type (upper) and Vpac2 receptor mutant (lower) SCN slices. Each line corresponds to an individual cell; red represents high and green, low levels of gene expression. Note the pronounced synchrony in wild-type SCN and its absence in the mutant slice. Reproduced with permission from Elsevier Ltd @ 2006. Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H & Hastings MH 2006 Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Current Biology 16 599–605.
Importantly, these imaging techniques have revealed novel aspects of the SCN as a circuit of oscillatory cells. First, although synchrony between cells is very tight in the SCN, they do not share completely common phases. In long-term slice cultures, there is a spatial order such that gene expression occurs first in the dorsomedial SCN shell and then progresses in a wave, ventrally and laterally (Yamaguchi et al. 2003). Second, in acute slices, the entraining effect of previous lighting schedules is apparent and SCN cells can be seen to segregate into distinct phase clusters based upon the peak time of their circadian gene expression rhythm (Quintero et al. 2003). This photic control over the distribution of cellular phases may well underlie photoperiodic time measurement, with longer days of summer represented by a more extensive phase distribution in the SCN and a compressed nocturnal melatonin profile in the pineal. Continuous light has an even more pronounced effect, causing the molecular rhythms of SCN neurons to become asynchronous across the slice, an effect that underlies the loss of behavioural rhythms under such conditions (Ohta et al. 2005).
Intercellular signalling is also important in maintaining as well as synchronising the cellular clocks. If it is interrupted by blockade of action potentials following the application of tetrodotoxin, molecular cycles within individual SCN cells not only desynchronise, but also lose amplitude (Yamaguchi et al. 2003, Maywood et al. 2007). An important mediator of this synchronisation and maintenance of molecular timekeeping is VIP, the neuropeptide of the SCN core. In SCN from mice lacking VIP or the Vpac2 receptor for VIP, the amplitude of circadian gene expression is dramatically reduced and interneuronal synchrony is lost (Fig. 3⇑; Maywood et al. 2006b). As a consequence of this molecular disorganisation, the rhythms of electrical firing in the SCN and the circadian behaviour of the mice are compromised (Harmar et al. 2002, Aton et al. 2005). The loss of SCN competence in Vpac2 null mice leads to disrupted corticosteroid and oestrous rhythms, which in turn are accompanied by changes in circadian gene expression in peripheral tissues (Dolatshad et al. 2006, Sheward et al. 2007). The interdependence of the core molecular loop and SCN neural circuit activity is illustrated further in mice lacking either Cry1 or Per1. When the SCN circuitry is intact, cellular rhythms are maintained despite the absence of one or other factor (as are behavioural rhythms). When the SCN neurons are dispersed into culture, however, their molecular timekeeping is compromised, by loss of either Cry1 or Per demonstrating that intercellular coupling enhances the robustness of cellular oscillators as well as synchronising them (Liu et al. 2007a). These observations raise intriguing question about the functional architecture of the SCN and its clock cells: what are the linkages between electrical activity and molecular timekeeping (Nitabach et al. 2005), what is the nature of the interactions between amplitude and synchrony of cellular oscillators and how do the core and shell of the SCN interact?
Outputs from the SCN and the signalling of circadian time
Neural projections from the SCN are well placed to drive endocrine and other circadian cycles, albeit via indirect poly synaptic connections (Buijs & Kalsbeek 2001). The principal circadian relay is a crescent-shaped continuum of the medial hypothalamus, running from the sub-paraventricular zone adjacent to the SCN, dorsally and caudally into the dorsomedial hypothalamus (Saper et al. 2005). Extensive links therefrom feed into arousal and sleep-regulatory centres of the orexinergic system and ventrolateral preoptic area respectively, plus a diversity of autonomic centres. Consequently, most viscera receive SCN-dependent circadian time cues via their parasympathetic and/or sympathetic innervation (Kalsbeek et al. 2006). In the adrenal, for example, light and circadian signals from the SCN can drive glucocorticoid (GC) synthesis and release without accompanying hypothalamo-adenohypophysial activation (Ishida et al. 2005). Similarly, nocturnal secretion of melatonin, an important regulator of seasonal rhythms in many species (Hastings 1991) and sleep efficiency in humans (Wyatt et al. 2006), is driven by a multi-synaptic pathway extending from the medial hypothalamus to the sympathetic afferents of the pineal gland. Within the hypothalamus, projections from the SCN to the paraventricular nucleus, the preoptic area and the medio-basal nuclei are able to regulate daily rhythms of adrenocorticotrophins (ACTH), gonadotrophins and metabolic hormones respectively (Kalsbeek et al. 2006).
The neurochemical nature of the SCN signals is of current interest, not least because neuronal transplantation studies have indicated that diffusible paracrine factors may play a central role. A screen for secreted factors of the suprachiasmatic nucleus (Kramer et al. 2005) has identified transforming growth factor-αand the neuropeptide cardiotrophin-like cytokine (Kraves & Weitz 2006) as likely mediators of SCN output. More recently, loss of the genes encoding either the SCN peptide prokineticin 2 (Prok2; Li et al. 2006, Hu et al. 2007) or its cognate receptor Prok2R (Prosser et al. 2007) has been shown to compromise circadian endocrine, behavioural and thermoregulatory rhythms, with particular effects in early night. Moreover, homeostatic regulation of sleep is compromised in mice lacking the Prok2 gene. Loss of the receptor, which is highly expressed in SCN target areas including the medial thalamus and dorsomedial hypothalamus, or ligand had no effect however upon cellular timekeeping in the SCN itself, even though both are expressed at high levels in the nucleus. It seems likely therefore that the SCN secretes a sequence of distinct ‘time-stamped’ factors and is thereby able to define serial phases of the circadian cycle within its diverse target sites.
Circadian clocks in peripheral tissues
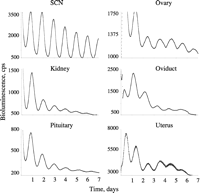
Following their discovery, clock genes were shown to be expressed in many peripheral tissues and in a circadian fashion. The conventional viewpoint was that these cycles were passively driven by efferent SCN signals: endocrine, autonomic and behavioural (e.g. rhythms offeeding and digestion). This view was overturned by the demonstration that mammalian fibroblasts continue to express circadian cycles of clock gene expression in long-term culture following a serum shock (Balsalobre et al. 1998). Real-time imaging of primary tissues cultured from bioluminescent reporter animals extended this to show that most major organ systems (including liver, heart kidney and skeletal muscle) contain intrinsic circadian clocks (Yamazaki et al. 2000; Fig. 4⇓). One reason to think that these peripheral clocks are less competent than the SCN is that with prolonged culture the rhythms of slice or cell cultures dampen, whereas SCN rhythms are sustained indefinitely. Single cell imaging of fibroblast cultures has shown, however, that the individual cells continue to express high-amplitude circadian cycles but lacking any means of synchronisation, the cells drift out of phase and aggregate bioluminescence or fluorescence rhythms across the culture lose definition (Nagoshi et al. 2004, Welsh et al. 2004). This suggests that the clocks of peripheral cells are as good as those of the SCN, and in general peripheral clocks use essentially the same molecular components as the SCN and show the same effects of circadian mutations (Yagita et al. 2001, Oster et al. 2006, Godinho et al. 2007). There are, however, differences between them. First, Clock is essential for the peripheral oscillators to function (DeBruyne et al. 2007a,b) whereas it is not for the SCN, where Npas2 can compensate for its absence. Equally, the clockwork of peripheral tissues (but not SCN) cannot function in the absence of Cry1 or Per1, and in dispersed fibroblast cultures Per2 is also essential (Liu et al. 2007a). Intriguingly, if SCN neurons are dispersed they too become dependent upon Per1 and Cry1. What makes the SCN clock cells different from other cell types, therefore, is their ability to both synchronise and sustain each other via interneuronal, circuit interactions. But paradoxically, their dependence on circuit inputs means that when intercellular signalling is compromised, as in VIP mutants, the cellular oscillators lose competence, becoming less effective than asynchronous, dispersed fibroblasts.
Circadian timekeeping in peripheral tissues. Representative plots of bioluminescence from organotypic slices of various tissues harvested from PER2::LUC reporter mouse and recorded in static culture using a photomultiplier assembly. The SCN sustain high-amplitude cycles throughout the recording. Peripheral tissues are also highly rhythmic, but as recording continues the amplitude and precision of the cycles decline, the rates varying between different organs. These declines are considered to arise from desynchrony of the individual cellular oscillators within each tissue. Acute activation of the tissues with serum shock or various pharmacological agents can re-synchronise the cells and restore high-amplitude cycles. In vivo, endocrine and other cues signalling SCN time will maintain and synchronise these peripheral clocks.
The view of the circadian axis is now a hierarchical one in which autonomous cellular oscillators distributed across the organism are synchronised by SCN-dependent cues (Fig. 5⇓), and this synchronisation sustains circadian organisation at the level of particular organs and also ensures appropriate internal synchronisation between different physiological and metabolic systems. An important question, therefore, is what maintains internal synchrony and what are the relative contributions to tissue rhythms of local clocks and rhythmic activation by systemic factors. DNA microarray studies indicate that 5–10% of local transcriptomes are under circadian regulation, whilst 10–20% of the proteome may be similarly rhythmic (Reddy et al. 2006), and many of these circadian targets are key elements of vital functions such as nitrogen or carbohydrate metabolism, control of oxidative pathways, cell division, etc. For example, the clock in the adrenal cortex drives a local transcriptome in which many genes involved in corticosteroid biosynthesis are under circadian regulation. Moreover, this local clock gates the response of the adrenal cortex to ACTH, itself a circadian cue dependent upon the SCN (Oster et al. 2006). By using local over-expression of Rev-erbαto suppress the clockwork in the liver of otherwise normal mice, Kornmann et al. (2007) showed that ca. 90% of the hepatic circadian transcriptome depends upon the local clockwork for its oscillation. That is to say, rhythmic systemic cues cannot sustain the bulk of tissue circadian rhythms if the local clock machinery is compromised.
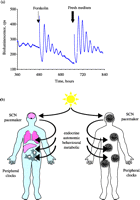
Synchronisation of circadian timekeeping across the body. Synchronous circadian gene expression in cultures of NIH3T3 fibroblasts transfected with a Per2::luciferase reporter construct can be initiated by addition of forskolin or fresh culture medium. Following acute activation, individual cells remain rhythmic but lose synchrony and so bioluminescence rhythms across the culture dampen. Lower panel presents schematic view of circadian co-ordination across the individual in which the primary pacemaker of the SCN, entrained to solar time by retinal afferents, maintains and synchronises tissue-based clocks in the major organ systems by a blend of endocrine, autonomic and behavioural (feeding-related) cues. Redrawn from Maywood et al. (2006a). Reproduced with permission from Elsevier Ltd @ 2006. Maywood ES, O’Neill J, Wong GK, Reddy AB & Hastings MH 2006 Circadian timing in health and disease. Progress in Brain Research 153 53–269.
Intriguingly, one of the few genes that continues to cycle in the ‘clockless’ liver is Per2, highlighting it as a potential sensor of rhythmic cues and mediator of internal synchronisation, in the same way that retinally induced Per1 and Per2 are the entry points to resetting the SCN clockwork. The CRE sequences of these genes may therefore be critical for synchronisation of peripheral tissues and cells, and indeed activation of cAMP/CRE signalling by treatment with forskolin is a very potent inducer of circadian gene expression in culture. In vivo, however, multiple mechanisms likely subserve internal synchronisation, including feeding schedules (Stokkan et al. 2001), body temperature (Brown et al. 2002), autonomic pathways (Terazono et al. 2003) and hormones (Hastings et al. 2003). Pre-eminent amongst endocrine signals are the corticosteroids. Both in vitro and in vivo, acute activation of GC receptor signalling can phase shift circadian timekeeping in cells and tissues (Balsalobre et al. 2000), although the SCN are unaffected by such signals, as they lacking the receptor. In fact, rhythmic secretion of GC seems to be a critical mediator of SCN output and can oppose the resetting actions of other cues, e.g. feeding (Le Minh et al. 2001). It is not too fanciful to view the SCN–adrenal–GC-axis as the body’s time standard, and it is remarkably potent. A single treatment of SCN-ablated mice with dexamethasone can activate ca. 60% of the liver circadian transcriptome, including core clock and clock-controlled genes that carry GC-response elements (GREs; Reddy et al. 2007). Amongst these are transcriptional regulators such as Dbp, hepatocyte nuclear factor 4α(Hnf4 α) which in turn direct the expression of their own target genes (for example, ornithine transcarbamylase, cytochrome P450 enzymes and carboxylesterases) many of which do not carry GREs but are expressed rhythmically and are critical tovarious vital processes such as ureagenesis, detoxification, nutrient and drug metabolism (Gachon et al. 2006). In addition, circadian control over transcriptional co-activators such as PGC1α(Liu et al. 2007b) will further direct the effects of transcriptional regulators such as Hnf4α(Puigserver 2005) and the response of metabolic control genes to both indirect and direct GC activation respectively (Jang et al. 2007). In this way, the gamut of hepatic physiology is co-ordinated in time and matched to the alternating demands of the SCN-dependent daily rest/activity cycle.
Conclusion: circadian clocks and health
Recognition of the temporal dynamics of physiology, its underlying control by cycles of gene expression and the role of hormones in synchronising these programmes within and between tissues brings a completely new perspective on the relationships between circadian clocks, health and disease. As a consequence of its evolution, the human body functions optimally when its component parts follow coherent daily cycles, in tune with each other and with solar and social rhythms. Consequently, these natural daily variations can aggravate, in a time-dependent way, pre-existing pathologies such that, for example, cardiovascular and cerebrovascular crises are most common in the hours after waking when the clock up-regulates cardiovascular activity in preparation for a new day (Hastings et al. 2003). In addition, when this internal coherence is disturbed by genetic disorder, the demands of modern working schedules or experimental manipulations, there are long-term consequences for health. For example, disruption of this regular endocrine programme by poor sleep patterns can have a severe impact on metabolic and mental health (Van Cauter et al. 2007). Some of these effects may appear to be non-specific; for example, increased mortality of aged mice arising from chronic ‘jet-lag’ (Davidson et al. 2006a) or metabolic syndrome in Clock mutant mice (Turek et al. 2005). On the other hand, animal models very clearly show that circadian mechanisms regulate essential physiological processes, including tissue growth (Fu et al. 2005), the control of blood pressure and heart rate (Curtis et al. 2007) and of blood glucose levels (Rudic et al. 2004). Clearly, the consequences of circadian disturbance for metabolic health will therefore be multi-factorial, but are nevertheless presented as greater risks of metabolic syndrome, hypertension and gastrointestinal disturbances in shift workers (Knutsson 2003, Oishi et al. 2005, Sookoian et al. 2007). With the identification of the genetic and molecular bases to circadian timekeeping, these long-term morbidities can now be readily understood by considering the likely effects of circadian disturbances on the expression of clock-controlled genes regulating the cardiovascular system, or the control of lipid and carbohydrate metabolism.
A more focussed involvement of the clock in disease comes within cancer. Rotating shift workers have a higher risk of developing cancers (Schernhammer et al. 2003, 2006), whilst in mice ablation of the SCN or rotating lighting schedules facilitate tumour progression (Filipski et al. 2002, 2004). In addition, Per2 mutation renders mice more prone to spontaneous and induced tumours (Fu et al. 2002) whilst over- or under-expression of Per1 can reduce or increase the rate of apoptosis in human cancercells (Gery et al. 2006). In vivo, a disrupted clock may influence tumour development in several indirect ways, for example, by altering immunocompetence, growth factor expression or the endocrine environment. Critically, the cycle of cell division is highly circadian in proliferating tissues such as oral mucosa and skin (Bjarnason et al. 2001), and by restricting when competent cells might divide to a narrow temporal window, the circadian timing system has potentially oncostatic effects. This becomes particularly relevant with the discovery that tumour cells also possess a functional clockwork (Filipski et al. 2004, Davidson et al. 2006b) that is entrained to factors within the host. Consequently, a disrupted host clock may open this gate within the tumour, thereby allowing increased proliferation. A direct molecular mechanism for such effects lies in the identification of several cell cycle regulators as clock controlled genes, including Wee-1, several cyclins and c-myc (Fu et al. 2002, Matsuo et al. 2003, Reddy et al. 2005). In a clinical context, these interactions are open to exploitation by seeking to time the delivery of cytotoxic therapies to coincide with points of vulnerability within tumour cells (chronotherapy) but such regimes also need to consider the effect of circadian processes on drug metabolism and inactivation (Levi & Schibler 2007) if efficacy is to be maximised. In short, therefore, knowledge gained over the last decade of the molecular and cellular bases of biological clocks has brought circadian time to the forefront of physiology, and thereby shown how temporal disorganisation can underlie major systemic illness. Indeed, the hidden but accruing costs of rotational shift work are set to become a major issue in public health. Conversely, these revelations also offer unanticipated opportunities for the development of targeted therapies, which will utilise central and local clockworks to manage and exploit circadian vulnerabilities in disease.
Acknowledgments
The authors’ work is supported by the Medical Research Council, the Biotechnology and Biological Sciences Research Council and the FP6 EUCLOCK programme. Drs Beth Klerman, Chuck Czeisler and Steve Lockley (Harvard Medical School) generously assisted in the creation of Fig. 1⇑, and Mr Paul Margiotta (MRC LMB) provided excellent assistance with illustrations. The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
- Received in final form 10 September 2007
- Accepted 12 September 2007
- Made available online as an Accepted Preprint 12 September 2007
- Society for Endocrinology