Hormonology: a genomic perspective on hormonal research
- Division of Reproductive Biology, Department of Obstetrics and Gynecology, Stanford University School of Medicine, 300 Pasteur Drive, Stanford, California, 94305-5317, USA
- 1Endocrinology Unit and EA 1533, University Pierre et Marie Curie, Genetics of Human Reproduction, Hôpital Saint-Antoine, 184 rue de Fg St Antoine, 75012, Paris, France
- (Requests for offprints should be addressed to A Hsueh; Email: aaron.hsueh{at}stanford.edu)
Abstract
Recent advances in comparative genomics allow a new paradigm for hormonal research. At the centennial of the first use of the term hormone by Ernest Starling, we reflected on the changing approaches in elucidating hormonal signaling mechanisms and highlighted the inadequacy of the term endocrinology, implying remote activation, to describe the diverse modes of hormone actions. Several examples were presented to underscore the power of comparative genomics in the identification of new polypeptide hormones, receptors, and signaling pathways. We propose the use of the term hormonology to more accurately reflect the expanding boundaries of the discipline.
Introduction
In 1905, Ernest Starling published four landmark Croonian lectures, entitled The chemical correlation of the functions of the body. Dr Starling used the term hormone for the first time in his initial lecture, The chemical control of the functions of the body (Starling 1905). He discussed ‘. . . substances produced for effecting the correlation of organs within the body’ and indicated that ‘These chemical messengers, or hormones, have to be carried from the organ where they are produced to the organ which they affect by means of the blood stream.’ The term he applied, hormone (from the Greek, hormōn, meaning I excite or arouse) was suggested to him by William B Hardy, who had consulted with his colleague in classics at Caius College, Cambridge, W T Vesey (Rolleston 1936, Sawin 1969, Henderson 2005).
Prior to and parallel to the usage of hormones by Starling, the term endocrine was also gaining popularity. In 1855 Claude Bernard distinguished the products of the ‘ductless glands’ from other glandular products by the term ‘internal secretions’. Although the controversial self-injection of testicular extracts by Brown-Sequard in 1889 jumpstarted the field of experimental endocrinology, contemporaries of Starling dealt mainly with animal experimentation based on surgical parabiosis, organ removal, tissue extracts, and auto-transplantation. These studies highlighted the important role of circulating hormones in intercellular signaling. The term endocrine (from the Greek endo, meaning internal, and krinein, to separate) was used by the French physiologist Edouard Laguesse in 1893 when describing the islets of the pancreas as endocrine cells (Fossati 2004). He was also the first to call these islet cells the islets of Langerhans, to celebrate the work of this great anatomist. In 1904, M A Limon, a French histologist, discussed the ‘endocrine’ nature of interstitial cells (Limon 1904, Medvei 1982). Subsequently, the Italian physician Nicola Pende popularized the use of this term in his treatise Endocrinologie. Patologia Clinica degli organi a secrezione interna in 1915 (Pende and Antognetti 1915, Vague 1971). In 1917, the Association for the Study of Internal Secretions was established in the United States and the first issue of the journal Endocrinology was published. In 1946, the British Society for Endocrinology was founded. It was not until 1952 that the official name of the American organization was also changed to The Endocrine Society.
During the second half of the twentieth century, tissue cultures and genetic analyses were introduced to the field of hormone research and the classic endocrine actions of hormones were extended to include paracrine and juxtacrine actions for insulin-like growth factor I (IGF-I) and other factors. In some cases, intracrine actions of hormones were found wherein molecules act upon the same cells in which they were synthesized (O’Malley 1989). In addition to select glandular tissues, hormones were identified in neurons, kidney, heart, and embryonic structures. One could conclude that virtually all cells of the body are hormone-producing. Even for Starling’s gastrointestinal hormone, secretin, paracrine functions in the brain and reproductive organs have been demonstrated (Sherwood et al. 2000, Chow et al. 2004).
With the development of immunoassays for hormones originated by Yalow and Berson (Yalow & Berson 1959), endocrinologists measured fluctuations of serum hormones, and endocrinology became one of the most quantitative branches of medicine. Analyses of dynamic changes in circulating hormones led to the recognition of feedback-control mechanisms, a concept that influenced other fields of biology (Umbarger 1992). Concomitant with advances in cell and molecular biology, studies on hormonal ligands, receptors, and the associated signal transduction mechanisms rapidly became integral parts of different branches of biomedicine, ranging from developmental and cell biology to clinical medicine. Paradoxically, following its own success, endocrinology is no longer distinguishable from cellular biology, neurobiology, immune regulation, or even intracellular signal transduction. Hansson and his colleagues questioned, ‘Is it still meaningful to speak of basic endocrinology, or are all aspects of this discipline now absorbed in the larger field of molecular cell biology?’ (Hansson et al. 1996). Likewise, J D Wilson asked whether endocrinology could survive as a discipline in the twenty-first century (Wilson 2000).
Recent progress in genome sequencing has heralded a new chapter in hormonal research by enabling the identification of numerous new hormones, receptors, and signaling molecules based on phylogeny (Hsu & Hsueh 2000, Shaaban & Benton 2001). An integrated understanding of the mechanisms of intercellular communication is emerging based on these genomic and evolutionary analyses (Leo et al. 2002). It is becoming clear that hormones regulate diverse plasma membrane receptors, nuclear transcriptional factors, and ion channels. Comparative genomic analyses indicated that the emergence of metazoan complexity during evolution is associated with an impressive diversification of ligands and receptors that allowed both redundancy and promiscuity (Escriva et al. 2000, Ben-Shlomo et al. 2003). During evolution, the paracrine mode of cellular communication, stemming from contact between adjacent cells, gave rise to the endocrine mode in which given hormones, produced in remote tissues, traveled by blood circulation to act on target cells. The hypothesized paracrine-to-endocrine transition is supported by the existence of protein ligands for the receptor tyrosine kinase superfamily including both paracrine and endocrine members (Ben-Shlomo et al. 2003). A genome-wide analysis of plasma membrane receptors also has revealed that paracrine and autocrine networks outnumber endocrine interactions. For the homeostatic maintenance of body functions, paracrine regulation is the common mechanism and is prevalent during embryonic development whereas endocrine regulation represents an extension.
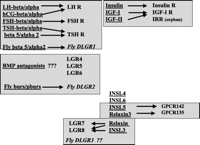
Two examples given below highlight the use of comparative genomics to understand hormonal signaling mechanisms. The first example deals with the identification of novel ligand and receptor genes similar to those of human glycoprotein hormones and their receptors. As shown in Fig. 1⇓, five orphan G-protein-coupled receptors have been identified in the human genome based on their sequence homology to human luteinizing hormone (LH), follicle-stimulating hormone (FSH), and thyroid-stimulating hormone (TSH) receptors and named as leucine-rich-repeat-containing, G-protein-coupled receptors (LGRs; Hsu et al. 1998, 2000). These receptors can be divided into two subgroups based on unique sequence features. Furthermore, all three types of LGR are present in the Drosophila genome with fly DLGR1, 2, and 3 corresponding to human LH/FSH/TSH receptors, human LGR4/5/6, and human LGR7/8, respectively.
To identify the ligands for the orphan human LGRs, we isolated two new glycoprotein hormone subunit genes in the human genome and named them α2 and β5. We demonstrated that α2 and β5 subunits are expressed in the same cells of the pituitary and could form heterodimers. Although the α2β5 heterodimers did not activate the newly identified orphan LGRs, this new hormone activated human TSH receptors and was named thyrostimulin (Nakabayashi et al. 2002). Furthermore, we identified in the fly genome orthologs to human α2 and β5 and demonstrated that fly α2 and β5 also formed heterodimers to activate the fly DLGR1 (Sudo et al. 2005; Fig. 1⇓).
Because there are no other human α- or β-like genes in the fully sequenced human genome, we hypothesized that hormones dissimilar to the glycoprotein hormone α and β subunits are ligands for the five orphan LGRs. Based on the evolutionary conservation of key residues between the LH receptor and LGR7/8, we demonstrated that gain-of-function point mutations in the human LH receptor responsible for male-limited precocious puberty could be used as the basis to generate constitutively active LGR7 and LGR8 (Hsu et al. 2002). These constitutive active LGR7 and LGR8 conferred cAMP stimulation, similar to the mutant LH receptors found in patients, indicating that LGR7 and LGR8 are likely to be activated by ligands capable of regulating cAMP production. Subsequent identification of the cognate ligands for LGR7 and LGR8 was made possible based on the common cryptorchid phenotypes of INSL3-null (Insulin-like 3) mice and mice with a large deletion of a genomic fragment containing LGR8 (Nef and Parada 1999, Zimmermann et al. 1999, Overbeek et al. 2001). Although INSL3 and related relaxin-like genes encode mature hormones with a two-chain structure similar to insulin and IGF-I, analyses of the human genome indicated that relaxin-like hormones are unlikely to interact with genes related to insulin receptors (Fig. 1⇓) because insulin and IGF receptors have cognate ligands and the orphan IRR (Insulin Receptor-like Receptor) could not be activated by relaxin. We, therefore, predicted that INSL3 and relaxin are the ligands for LGR7 and/or LGR8 and tested the ability of relaxin to activate cAMP production mediated by LGR7 or LGR8. Indeed, treatment with relaxin activated cAMP production in cells expressing either LGR7 or LGR8 (Hsu et al. 2002). Subsequent studies also demonstrated that INSL3 is the cognate ligand for LGR8 (Kumagai et al. 2002). In addition, recent studies demonstrated that the relaxin-like hormones INSL5 and relaxin3/INSL7 are capable of activating another group of G-protein-coupled receptors, GPCR142 and GPCR135 (Liu et al. 2005). It is likely that future derivation of bioactive ligands encoded by the remaining relaxin-like genes could allow further matching of the remaining orphan relaxin-like ligands with their receptors.
In contrast to the third group of orphan LGR7 and LGR8, no gain-of-function mutations could be generated for human LGR4, 5, or 6. Although recent studies using LGR4- and LGR5-null mice indicated the important roles of LGR4 and LGR5 genes in embryonic development and perinatal survival (Mazerbourg et al. 2004a, Morita et al. 2004), the ligands for these receptors are still unknown. Based on genomic analyses, we identified fly DLGR2 as the ortholog for human LGR4/5/6 and found this gene to be the receptor for bursicon, a neurohormone essential for tanning and sclerotization in diverse insects. Based on biochemical purification of the bursicon polypeptide and the tanning defects of the fly DLGR2 gene, an earlier study suggested that burs, a cystine-knot-containing polypeptide, is the cognate ligand for DLGR2 (Dewey et al. 2004). Because our preliminary data indicated that the recombinant burs polypeptide could not activate DLGR2, we searched the fly genome for additional cystine-knot-containing polypeptides and identified pburs (partner of burs). Interestingly, burs and pburs form heterodimers capable of activating the fly DLGR2 receptor (Luo et al. 2005), thus indicating that bursicon is a heterodimeric hormone. Because burs and pburs are homologous to human BMP antagonists, this study further raised the possibility that heterodimeric BMP (Bone Morphogenic Protein) antagonists act as candidate ligands for the orphan LGR4/5/6.
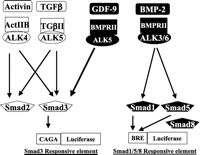
In addition to the identification of novel receptor and ligand genes to reveal new ligand–receptor pairs, a comparative genomic approach also facilitates identification of known receptors and downstream signaling mechanisms for orphan ligands. An example of this approach is the identification of GDF9 (Growth Differentiation Factor 9) receptors. GDF9 belongs to the large transforming growth factor β (TGFβ) family of genes with more than 30 members. As shown in Fig. 2⇓, several TGFβ/BMP ligands have been found to interact with type II and type I serine kinase receptors, leading to the activation of two sets of downstream Smad transcriptional factors. Although the oocyte-derived GDF9 stimulates granulosa cell proliferation, the exact receptors and signaling pathways for this ovarian paracrine factor were unclear. Instead of performing GDF9-binding assays and purifying the GDF9 receptors from granulosa cells for their identification, we hypothesized that GDF9, like other TGFβ/BMP genes in the same family, likely activates serine/threonine kinase receptors. However, searches of the human genome for genes with sequence homology to known type II and type I serine kinase receptor failed to reveal uncharacterized serine/threonine kinase receptor genes. Thus, one can hypothesize that GDF9 interacts with known type II and type I receptors in granulosa cells. Because there are only five type II and seven type I serine/threonine kinase receptors in the genome, we obtained the ectodomains of all type II receptors and demonstrated the ability of the BMP receptor II (BMPRII) ectodomain to block GDF9 stimulation of granulosa cell proliferation (Vitt et al. 2002). Likewise, we identified a GDF9-non-responsive cell line expressing BMPRII. In this cell line, overexpression of ALK5 (Activin Receptor-like kinase 5), but not the remaining six type I receptors, conferred GDF9 responsiveness (Mazerbourg et al. 2004b). The predicted role of BMPRII and ALK5 as type II and type I receptors for GDF9 was validated in granulosa cells following suppression of endogenous BMPRII and ALK5 expression using gene-knockdown approaches (Vitt et al. 2002, Mazerbourg et al. 2004b). The above examples indicated that evolutionary tracing of polypeptide ligands, receptors, and downstream signaling molecules in their respective ‘sub-genomes’ allows a new paradigm for hormonal research. Using a similar approach, receptors for three additional orphan GDP/BMP ligands have been recently identified (Mazerbourg et al. 2005).
In the postgenomic era it has become possible to integrate studies on endocrine hormones, growth factors, cytokines, gastrointestinal peptides, and neuropeptides, as well as immune factors, based on comparative genomic analyses of all human genes involved in hormonal signaling. In their original discovery of secretin (Bayliss & Starling 1902), Bayliss and Starling stated that ‘. . . secretin preparations made from the duodenum of the cat, rabbit, ox, monkey, man and frog are all active as regards to the pancreatic secretion of the dog.’ They concluded that, ‘the secretin of all these animals is one and the same body.’ At the centennial of the publication of the term hormone it is appropriate to reflect on the visionary deliberations of Starling and examine the potential applications of comparative genomic analyses of hormonal signaling mechanisms.
We propose hormonology, a term that more accurately reflects the scope of this field. The term endocrinology, implying remote activation, no longer describes the diverse modes of hormone actions. Indeed, Seymour Lieberman advocated, with apologies to the endocrine community, the use of hormonology in his 1986 Dale lecture to the British Endocrine Societies (Lieberman 1986). French scientists have continuously used the term hormonology to denote the biological aspects of the discipline, reserving the term endocrinology for the medical specialty. In 1997, Russian scientists also adopted this term (Kulinskii & Kolesnichenko 1997).
There is no doubt that endocrinology will survive as a discipline with sub-fields to emphasize a particular physiological focus such as growth, metabolism, reproduction, or bone metabolism. Starling stated at the end of his lecture that ‘. . . an extended knowledge of the hormones and their modes of action cannot fail to add largely to that complete control of the body which is the goal of medical science.’ At the beginning of the second century of hormone studies, the term hormonology would more accurately reflect the expanding boundaries of the discipline and our attempts to gain a complete knowledge of hormones and their modes of action.
Matching of several sets of paralogous polypeptide ligand and receptor genes in the human genome. Each set of ligand–receptor genes in the sub-genome is boxed to emphasize their evolutionary origins. R, receptor; hCG, human chorionic gonadotropin.
Diverse TGFβ family ligands interact with a limited number of type II and type I serine/threonine kinase receptors to activate two downstream Smad signaling pathways. The hypothesis that GDF9 likely interacts with one or more of the five type II and seven type I serine/threonine kinase receptors allows a focused search to reveal the role of BMPRII and ALK5 as GDF9 receptors, capable of activating the Smad2/3 pathway. CAGA, concensus nucleotide sequence in the promoter; BRE, BMP-responsive element.
Acknowledgments
The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
- Received 10 June 2005
- Accepted 12 June 2005
- © 2005 Society for Endocrinology